How to create an amino acid modifier in a scientific library in UNIFI - WKB5940
Article number: 5940
OBJECTIVE or GOAL
Create a custom amino acid modifier for intact protein or peptide mapping analysis in UNIFI.
ENVIRONMENT
- UNIFI
- waters_connect
PROCEDURE
The scientific library provides the most commonly used amino acid modifiers, but you can define new ones. To do so, from the Scientific Library section in the Administration area, select "Amino acid modifiers" from the Configure Scientific Library tab and follow the steps described below.
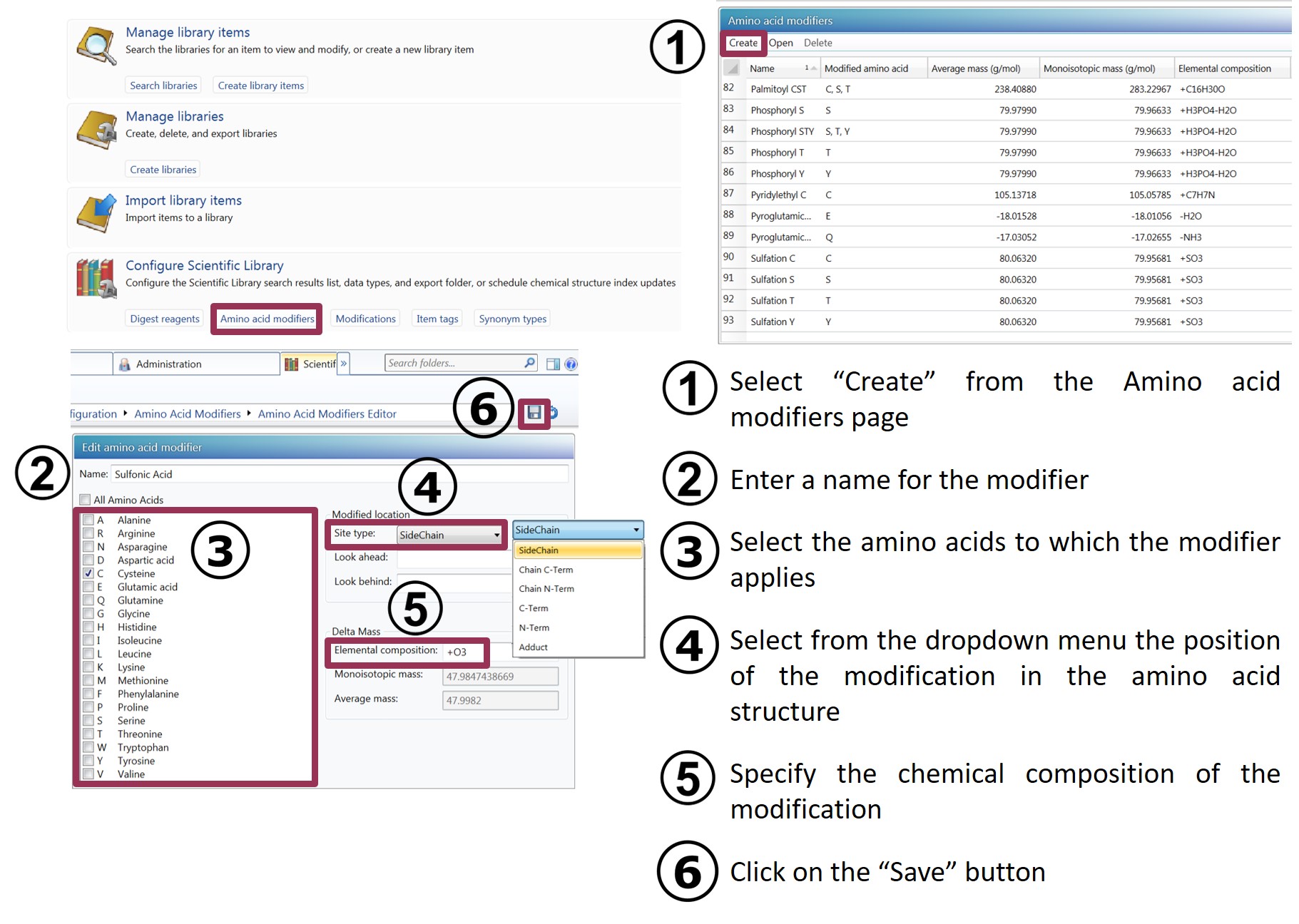
ADDITIONAL INFORMATION
- You can neither modify nor delete the predefined amino acid modifiers in UNIFI.
- This article is part of a series of articles on UNIFI scientific libraries (How to build and manage a scientific library in UNIFI - WKB190999) and can also be found with the following link: Advanced UNIFI: Scientific Library building and managing (715007127).
- Optionally, enter sequences for "look ahead" and "look behind". For a given amino acid sequence, "look ahead" is used to look for a matching sequence immediately following the residues to which the modifier applies. For a given amino acid sequence, "look behind" is used to look for a matching sequence immediately preceding the residues to which the modifier applies.
- Custom modifications defined with a look-behind sequence (either alone or in combination with look ahead sequence) are not applied correctly in analysis. See 118646.

