Custom amino acid modification is not applied to residue specified by look-ahead or look-behind settings - WKB118646
SYMPTOMS
- Despite a custom amino acid modification being applied with look-ahead and look-behind sequences to ensure that only a specific residue is modified in a particular protein sequence, other amino acids in the search results may be assigned the modification.
- For example, the "tail" of histone H3 contains eight lysine (K) residues, each of which may be acetylated. Modifying different residues has a different biological effect, and these lysines may also be subject to other modifications (see The Role of Histone Tails in the Nucleosome: A Computational Study, Erler et al., Biophysical Journal, 107, Vol 12, 2014).
- If you want to define an acetyl modification specific to K9, you could define a look-ahead sequence of "STG" and/or a look-behind sequence of "TAR" in the modifications editor of the UNIFI scientific library.
- With the example look-behind sequence, either in combination with a look-ahead sequence or on its own, the modification editor may show the modification specificity being applied correctly. However when this is used in an analysis method it will not be applied correctly in processing results that use that site-specific modification in the Amino Acid Modifications section of the Analysis Method.
- If you include only a look-ahead sequence, the modification editor shows the modification specificity being applied correctly, and it may also be applied correctly in processing results from an analysis method that uses that site-specific modification in the Amino Acid Modifications section of the method.
ENVIRONMENT
- waters_connect 3.2.0
- waters_connect 3.1.0
- waters_connect with UNIFI 3.0.0
- waters_connect with UNIFI 2.1.2
- waters_connect with UNIFI 1.9.13
- UNIFI 1.9_SR4
CAUSE
Incorrectly defined search term.
The look ahead and look behind functionality requires very specific syntax to be used in the search definition otherwise it won't work properly. Just typing in a sequence of amino acids is not enough. Unfortunately that syntax is not explained in the UNIFI help files (UNIFI), or in the Administration app help files (waters_connect)
FIX or WORKAROUND
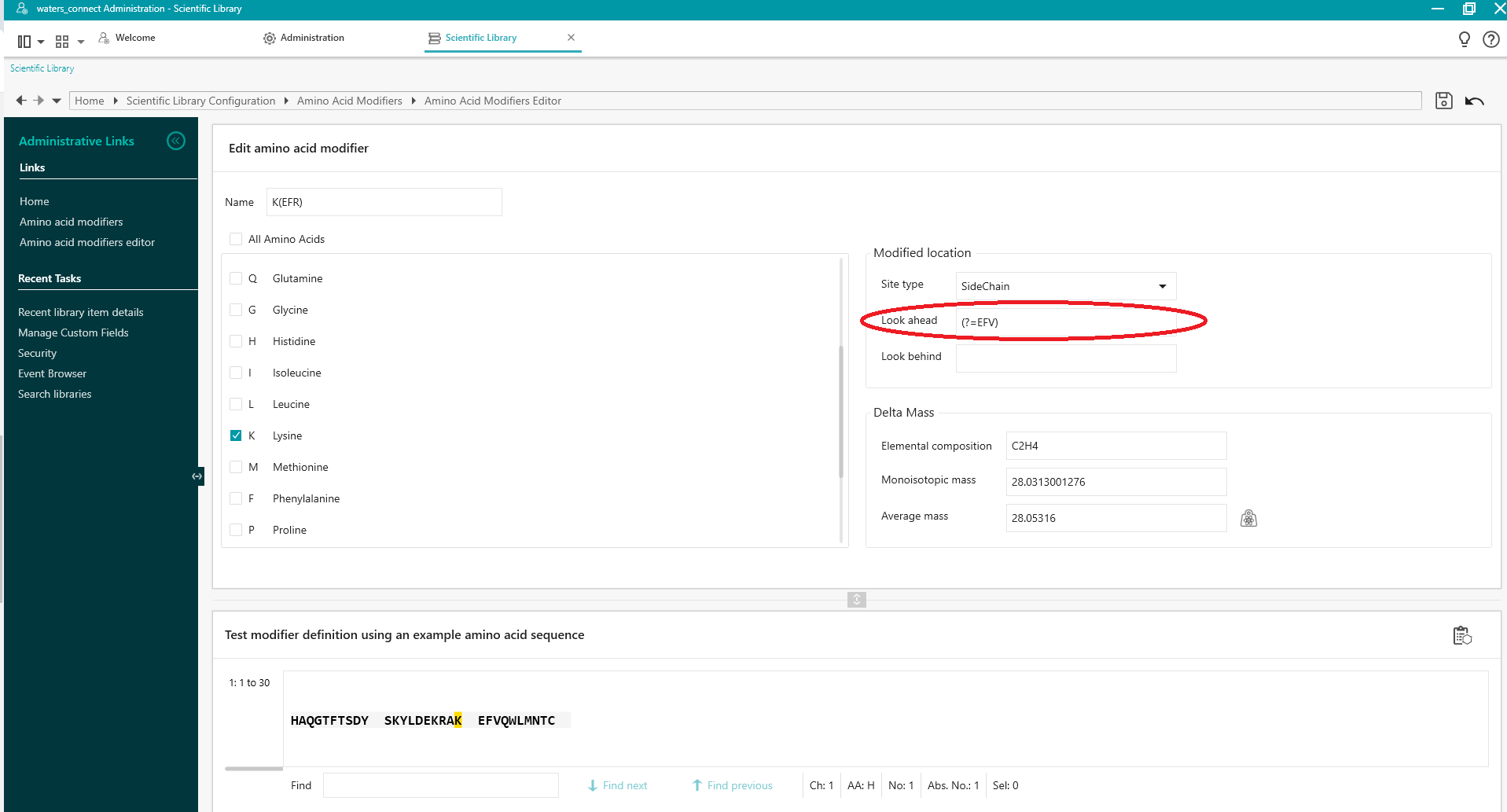
Look Ahead
To use a sequence of amino acids after the Applies To residue to target a modification at that specific residue, use the following syntax:
(?=pattern) where pattern is the sequence of amino acids.
For example, to use 'Look Ahead' to apply the defined modification only to K at position 20 in the sequence HAQGTFTSDYSKYLDEKRAKEFVQWLMNTC use (?=EFV) to search for K(?=EFV) (i.e. apply modification to K only if it is followed by EFV).
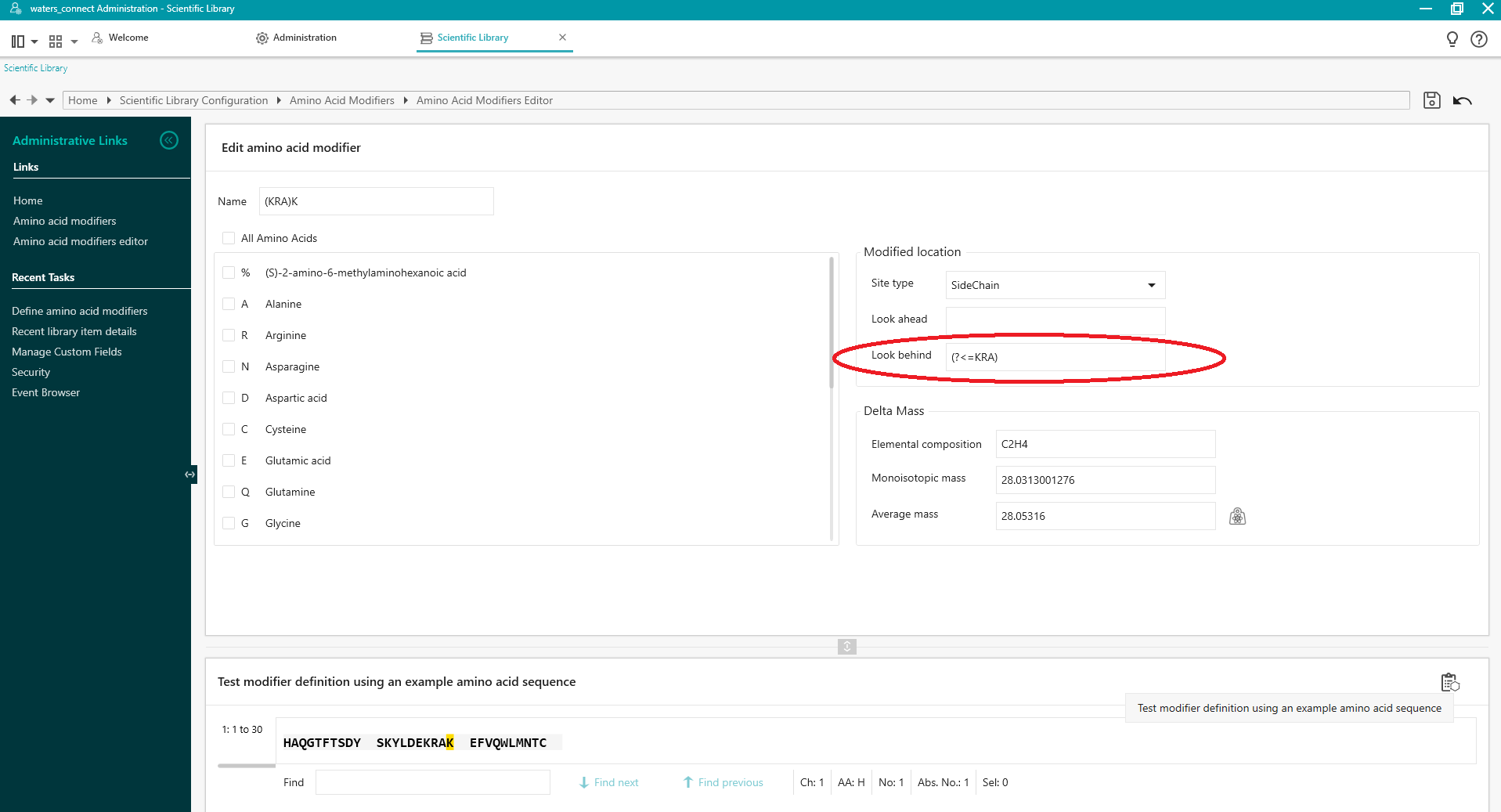
Look Behind
To use a sequence of amino acids before the Applies To residue to target a modification at a specific residue use the following syntax:
(?<=pattern) where pattern is the sequence of amino acids.
For example, to use 'Look Behind' to apply the defined modification to the K at position 20 in the sequence HAQGTFTSDYSKYLDEKRAKEFVQWLMNTC use (?<=KRA) to search for (?<=KRA)K (apply the modification to K only if it is preceded by KRA).
ADDITIONAL INFORMATION
id118646, SUPUNIFI, SUPWC, UNIFISVR, UNIFISW18, UNIFOPT, UNIFQLIC, UNIFSW17, UNIFSW18, UNIFSW19, UNIFWGLIC, UNIFWKLIC