Adduct error means m/z is wrong when peptide result is imported from a scientific library into a Tof 2D method - WKB54773
Article number: 54773
SYMPTOMS
- When a peptide is exported from results of a peptide mapping analysis to the scientific library and then imported to a Tof 2D method, the adduct used to calculate the expected m/z is wrong
- Export a peptide ID to Scientific Library:
- Expected m/z shown in library:
- Create 2D Tof method:
- When the Scientific Library entry is imported into the method, the expected m/z is wrong.
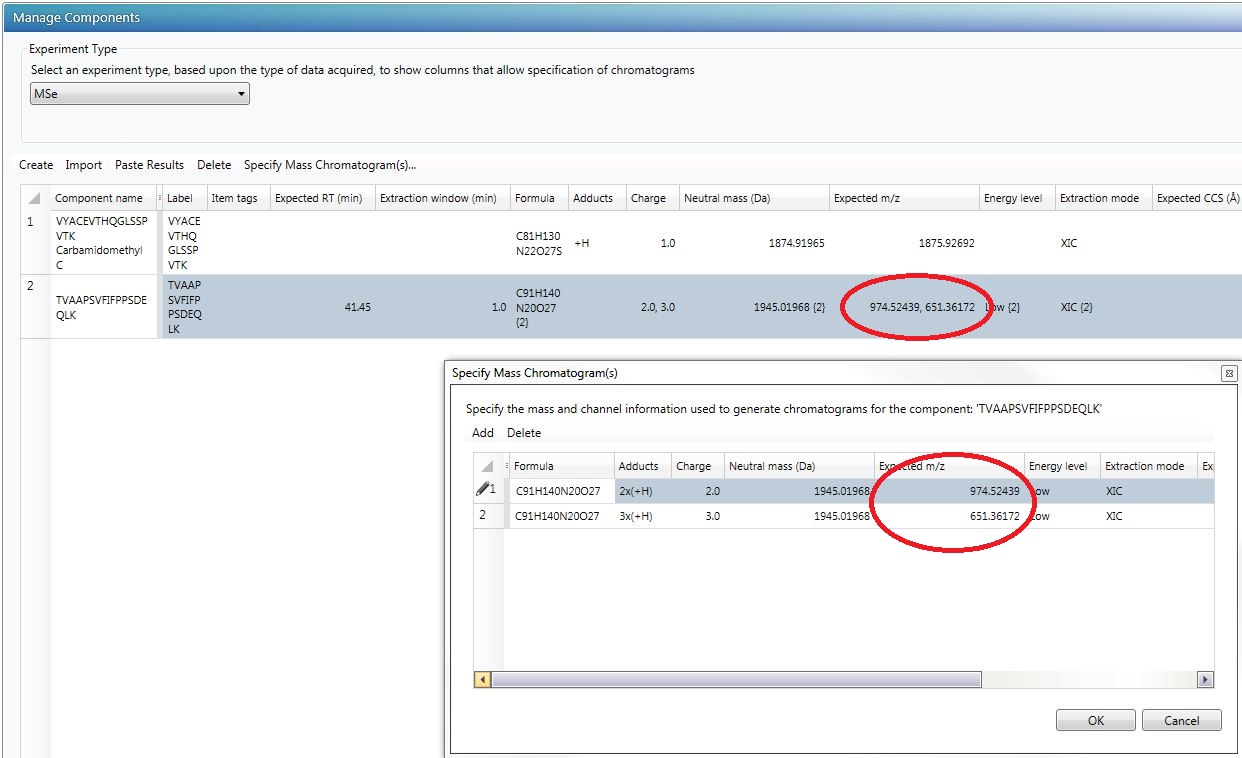
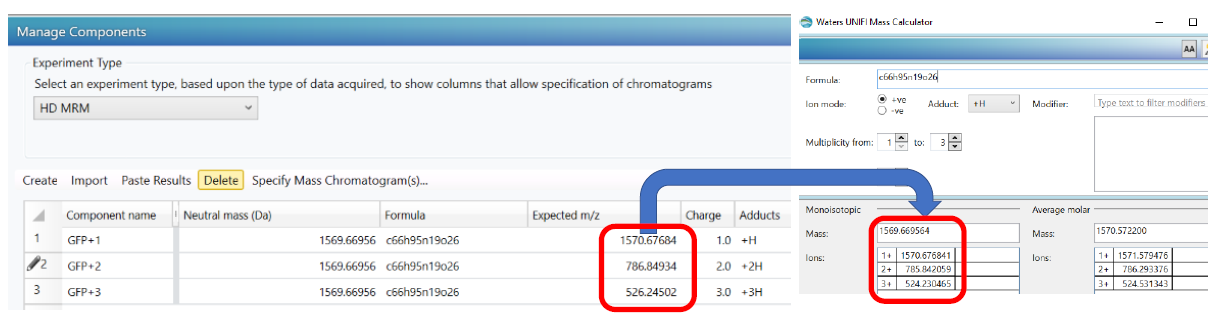
- Component masses calculated in the Manage Components section of Analysis Method > Purpose do not match the theoretical masses in the UNIFI Mass Calculator
ENVIRONMENT
- UNIFI 1.9 SR4
- UNIFI 1.9 SR3
CAUSE
Bug
(It seems the H+ mass is added twice before the neutral mass is divided by the charge to generate the m/z.
FIX or WORKAROUND
Create custom 2+ and 3+ (and other) adducts and manually select them in the adducts column of the "Specific Mass Chromatogram(s)" pane. This forces the method generator to redo the m/z calculation and get it right.
ADDITIONAL INFORMATION
Raised as fault INFULA-812.
According to the fault record, this problem is fixed in UNIFI 1.9.9 and later and did not occur in UNIFI 1.8.2.
id54773, SUPUNIFI, UNIFISVR, UNIFISW18, UNIFOPT, UNIFQLIC, UNIFSW17, UNIFSW18, UNIFSW19, UNIFWGLIC, UNIFWKLIC

